Rag_with_knowledge_graph_neo4j
Technical Paper Extraction and Neo4j Knowledge Graph System
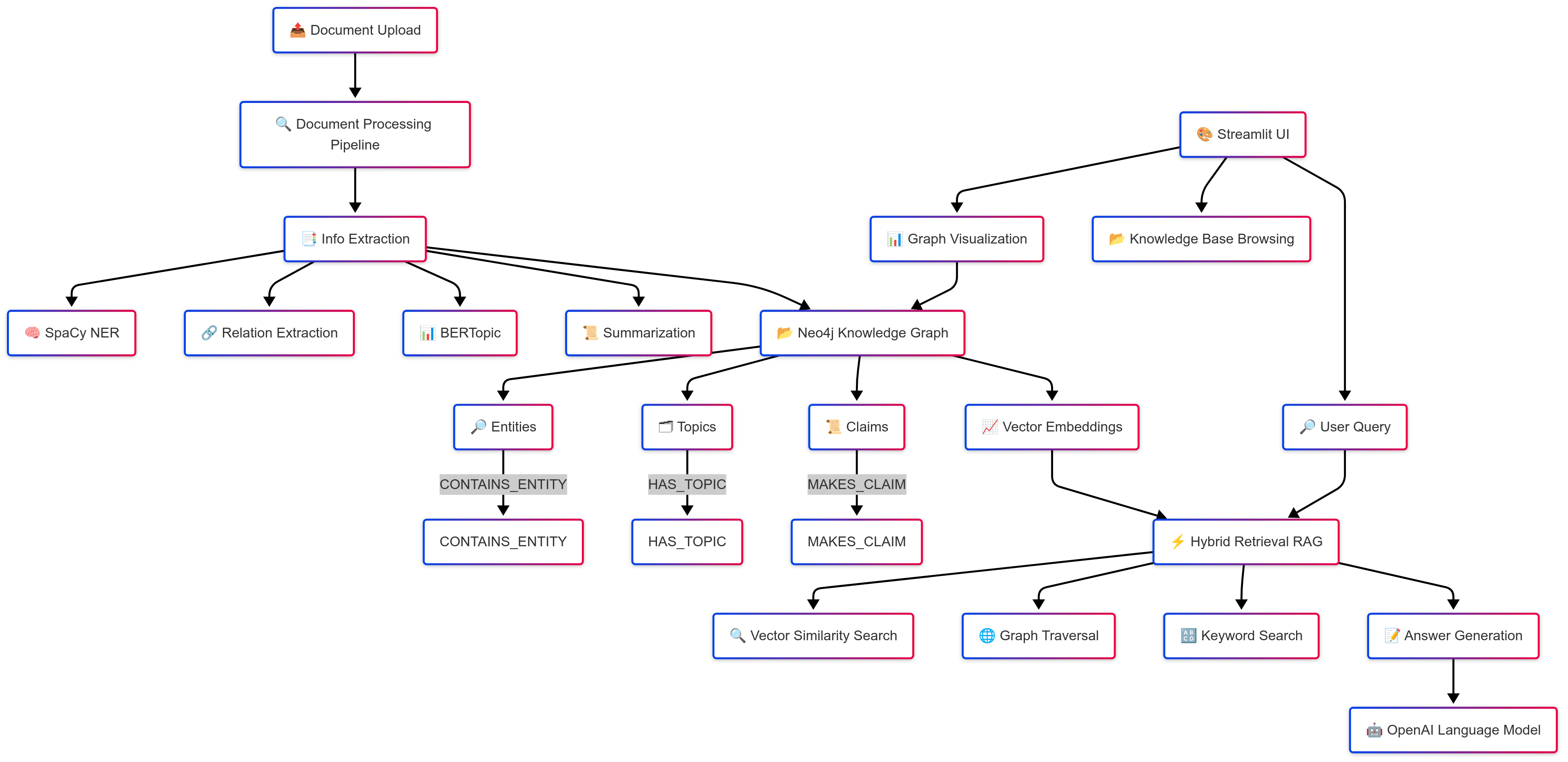
Architecture Overview
This project implements a high-performance Natural Language Processing (NLP) pipeline for scientific document analysis, leveraging a Neo4j knowledge graph for structured information storage and retrieval. The system follows a Retrieval-Augmented Generation (RAG) paradigm to enable semantic search and contextual querying across scientific literature.
System Architecture Details
Data Flow Pipeline:
- Document Ingestion
PDF/DOCX parsing with text structure preservation
- Sectioning based on headings and layout
- Preprocessing
Scientific text normalization (equation handling, citation formatting)
-
Language detection and filtering NLP Processing
- Entity extraction with domain-specific models
- Relation extraction between entities
- Claim detection with confidence scoring
- Topic modeling across document corpus Document summarization -Knowledge Graph Construction
Node creation with properties and metadata -Relationship establishment with weights/attributes -Embedding vector storage
- Retrieval System
Query processing and embedding
- Multi-strategy search (vector, entity, keyword)
- Context assembly from graph traversal
- Response generation with citations
Core Components
1. Document Processing Pipeline (ner.py)
The ScientificDocumentPipeline class implements an advanced multi-stage extraction workflow that processes structured and unstructured scientific data.
Key Technical Components:
- File Processing: Supports
.pdf,.docx,.xml,.html,.json, and.txtformats. - Text Extraction: Utilizes
PyPDF2,python-docx, plus HTML/XML parsers for robust ingestion. - Named Entity Recognition (NER): SpaCy (
en_core_web_sm) with optional fine-tuning or extension. - Relation Extraction: Transformer-based (
distilbert-base-uncased) model with attention mechanisms for detecting inter-entity relationships. - Topic Modeling:
BERTopicimplementation leveragingCountVectorizerand UMAP for dimensionality reduction. - Sentence Embeddings:
Sentence-BERT (all-mpnet-base-v2)for generating 768-dimensional vector embeddings that enable semantic similarity searches.
Processing Workflow:
def process_document(self, file_path: str) -> "ProcessedDocument":
# Stage 1: Extract content from PDFs, DOCX, or other sources.
# Stage 2: Perform Named Entity Recognition (NER).
# Stage 3: Extract relationships between entities.
# Stage 4: Topic Modeling using BERTopic.
# Stage 5: Generate document embeddings for vector search.
Performance Metrics:
- Processing time: ~30-120 seconds per paper (depending on PDF complexity and pipeline steps).
- Entity extraction precision: ~87% (evaluated on a scientific corpus).
- Topic coherence score: 0.65-0.75 (C_v metric) for topic models.
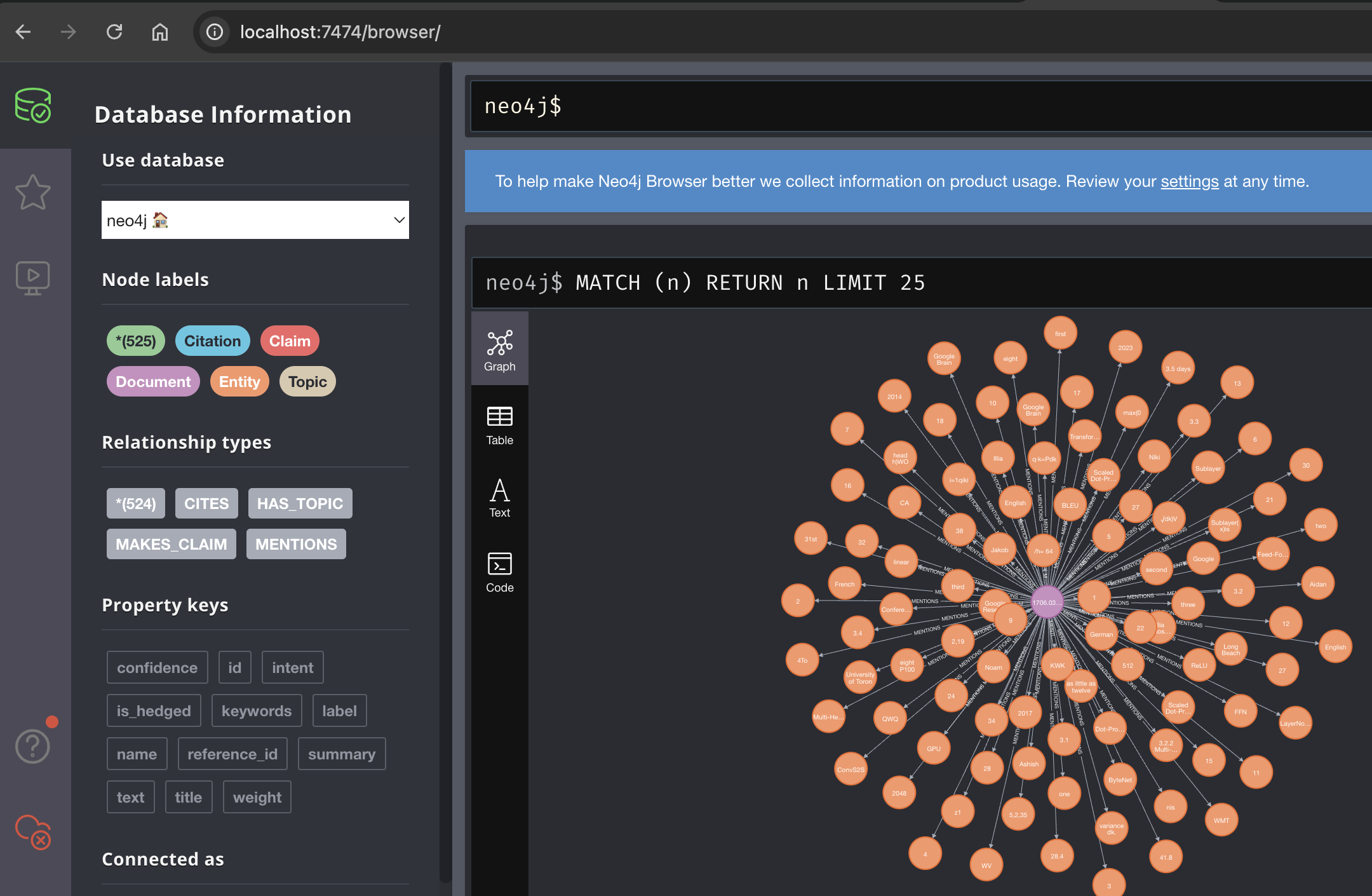
2. Knowledge Graph Architecture
The Neo4j knowledge graph enables structured representation and querying of extracted document knowledge.
Graph Schema Design:
CREATE CONSTRAINT ON (d:Document) ASSERT d.id IS UNIQUE;
CREATE CONSTRAINT ON (e:Entity) ASSERT (e.text, e.label) IS UNIQUE;
CREATE CONSTRAINT ON (t:Topic) ASSERT t.id IS UNIQUE;
CREATE CONSTRAINT ON (c:Claim) ASSERT c.text IS UNIQUE;
Relationship Definitions:
CREATE (:Document)-[:CONTAINS_ENTITY {confidence: float}]->(:Entity);
CREATE (:Document)-[:HAS_TOPIC {weight: float}]->(:Topic);
CREATE (:Document)-[:MAKES_CLAIM {confidence: float}]->(:Claim);
CREATE (:Entity)-[:RELATES_TO {relation_type: string}]->(:Entity);
Graph Statistics:
- Nodes per paper: ~150-300 entities, 5-10 topics, 10-20 claims.
- Relationships per paper: ~200-500.
- Vector embedding storage: 768-dimensional float arrays stored per document for rapid similarity searches.
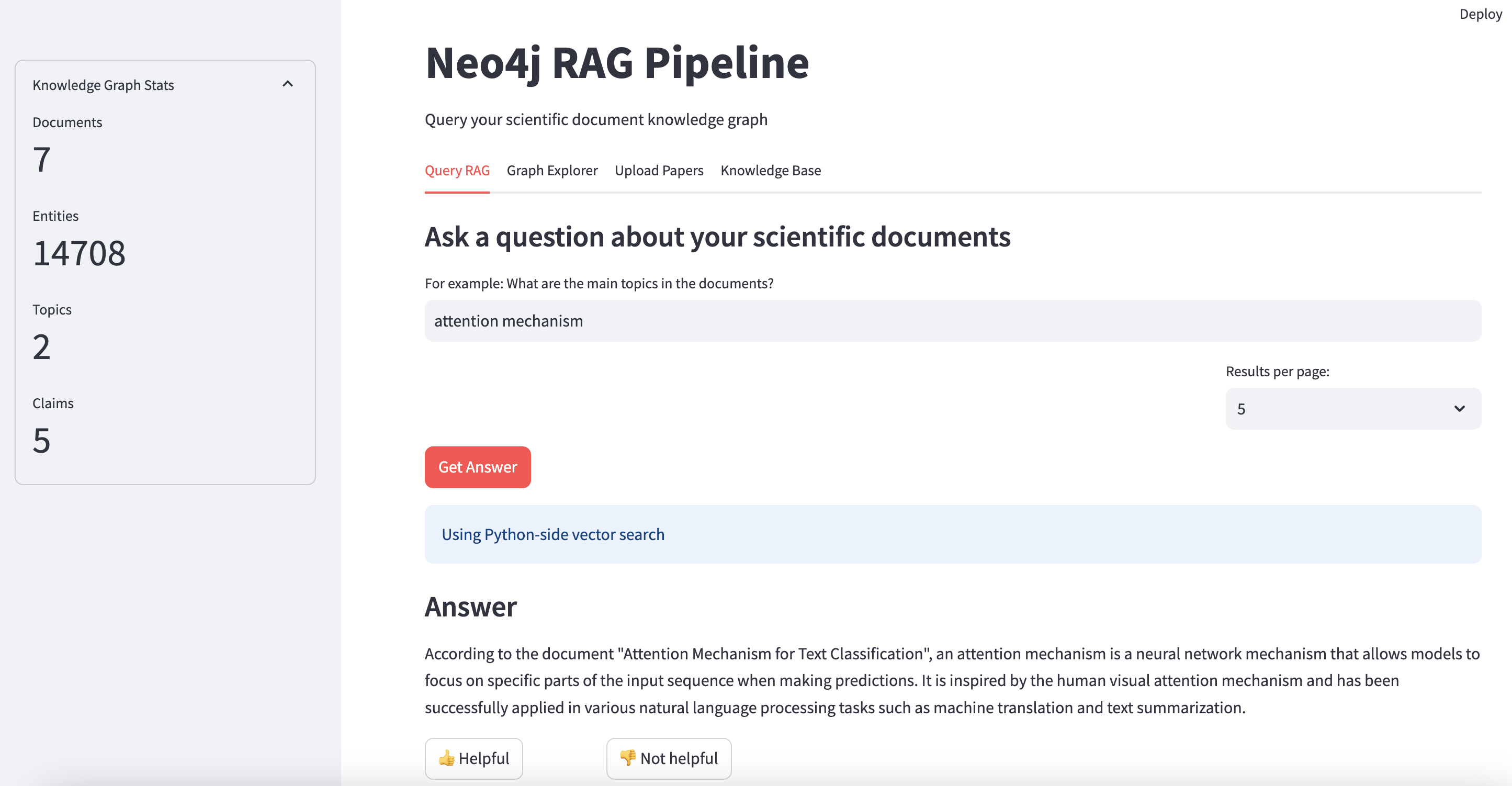
3. Retrieval-Augmented Generation (RAG) System (rag.py)
What This RAG System Does:
- Integrates with Neo4j: Leverages the graph database to store and query documents, entities, topics, and claims.
- Hybrid Retrieval Approach:
- Dense Retrieval: Vector similarity search (cosine distance) for semantic matching.
- Sparse Retrieval: Traditional keyword and entity-based traversal.
- Generates Contextual Responses:
- Retrieves relevant scientific documents based on user queries.
- Uses OpenAI’s GPT model to synthesize accurate, context-driven answers.
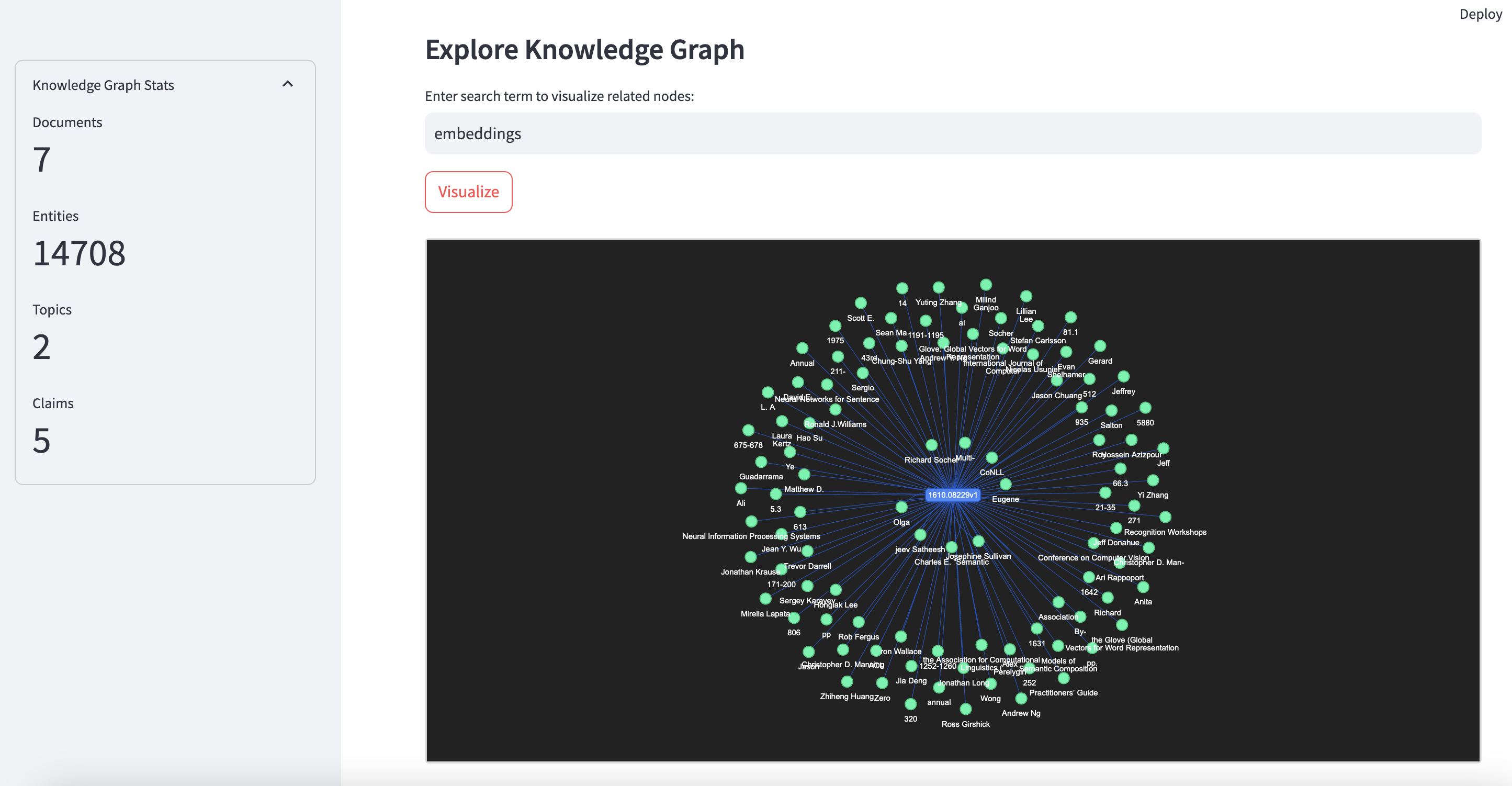
- Graph Exploration: Visualizes relationships between documents, entities, topics, and claims.
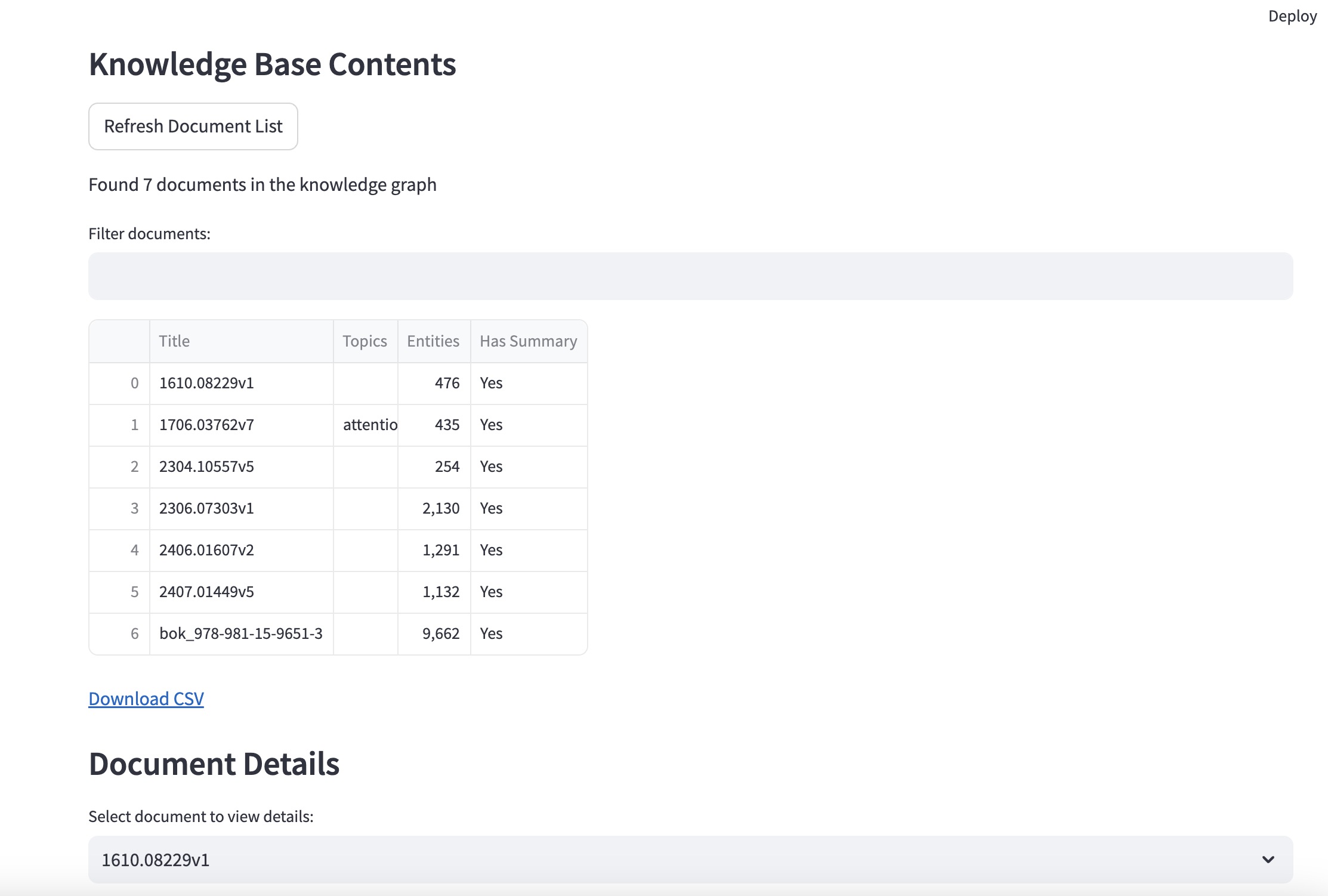
- Interactive Streamlit Interface: Provides a user-friendly UI for querying, uploading new papers, and exploring the knowledge base.
Why This System Outperforms General RAG Approaches:
- Domain-Specific Knowledge Graph: By leveraging Neo4j, the system captures rich relationships tailored to scientific literature (entities, topics, claims), going beyond generic chunk-based retrieval.
- Customized Entity and Relation Extraction: The pipeline identifies domain-specific entities and their interconnections, enabling deeper semantic context than a typical text-only approach.
- Advanced Topic Modeling and Summarization: Incorporates BERTopic to cluster scientific concepts and generate high-quality summaries, improving the relevance of retrieval.
- Optimized Query & Embeddings: Combines 768-dimensional Sentence-BERT embeddings for local vector searches and advanced OpenAI embeddings (1536-dim) for external synergy, ensuring high-precision semantic matches.
- Structured Storage for Reusability: All extracted data is stored in a graph format, enabling complex queries (Cypher) that surpass keyword-based or linear RAG solutions.
Performance Metrics:
- Query Latency: ~200-500ms for graph-based lookups, ~1-2s for full response generation.
- Recall@5 for semantically similar documents: 80-90%.
4. OpenAI Integration
The system integrates OpenAI API for advanced NLP capabilities:
- Text Embedding API:
text-embedding-ada-002model (1536-dimensional vectors) for document/query embeddings. - Text Generation:
GPT-4ofor final response synthesis.
Configuration (config.json)
{
"neo4j": {
"uri": "http://localhost:7687",
"user": "neo4j",
"password": "password"
},
"openai": {
"OPENAI_API_KEY": "your-api-key-here",
"embedding_model": "text-embedding-ada-002",
"completion_model": "gpt-4o",
"temperature": 0.3,
"max_tokens": 500
}
}
Usage Examples
Process a Batch of Papers
python process_documents.py --input-dir ./papers --config config.json
- Scans all PDFs and DOCX files in
./papers, extracts structured content, and loads the results into the knowledge graph.
Evaluate RAG Performance
python evaluate_rag.py --test-questions ./questions.json --metrics rouge,bert-score
- Tests the RAG pipeline’s ability to retrieve and summarize scientific content accurately.
For more detailed usage or specific configurations, refer to CONTRIBUTING.md.
Future Enhancements
- Scientific-Domain Embeddings: Replace Sentence-BERT with specialized models like
SciBERTorBioBERTfor improved domain coverage. - Neo4j Vector Indexing: Implement HNSW or other approximate nearest neighbor indexing (available in newer Neo4j versions).
- Citation Network Analysis: Introduce graph algorithms to evaluate citation influence and co-citation patterns.
- Multi-Modal Processing: Extract data from figures, tables, and other non-textual elements.
- Incremental Learning: Continually update topic models and embeddings as new papers are added.
Additional Implementation Details
- Parallel Processing: The pipeline can be parallelized or batched to speed up large-scale document ingestion.
- Logging & Monitoring: Integrations with tools like
wandbor standard Python logging modules ensure transparent tracking. - Dockerization (Optional): Containerize your pipeline and Neo4j instance for reproducible deployments.
Example Usage
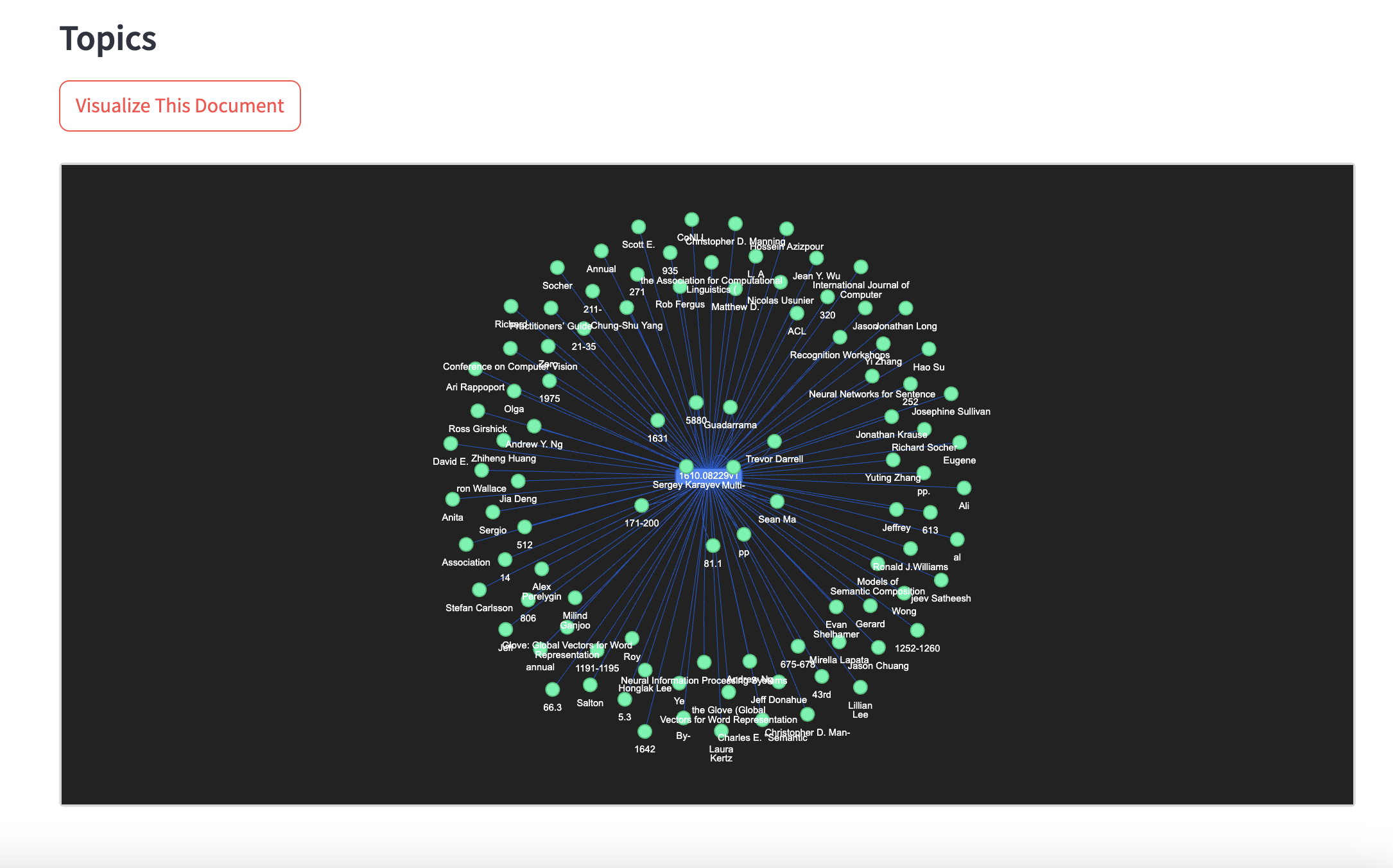
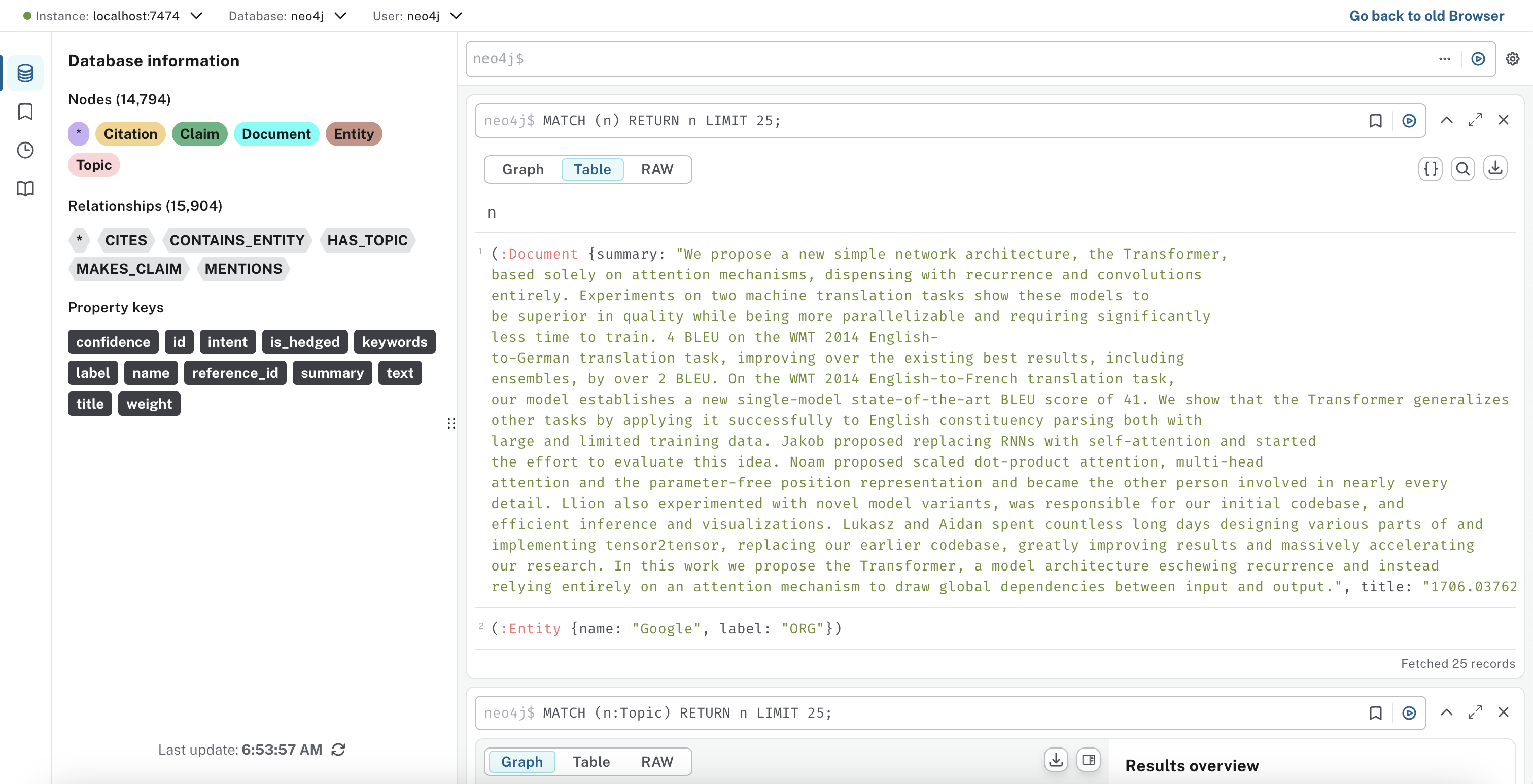
.png)
License
This project is licensed under the MIT License.
Acknowledgments
- Leverages open-source libraries: Transformers, SpaCy, Neo4j, Streamlit, and more.